
TopoGromacs: Automated Topology Conversion from CHARMM to GROMACS within VMD. - Abstract - Europe PMC

Tutorial: Modelling post-translational modified proteins with GROMACS – Dr Anthony Nash MRSC: computational chemistry and life sciences
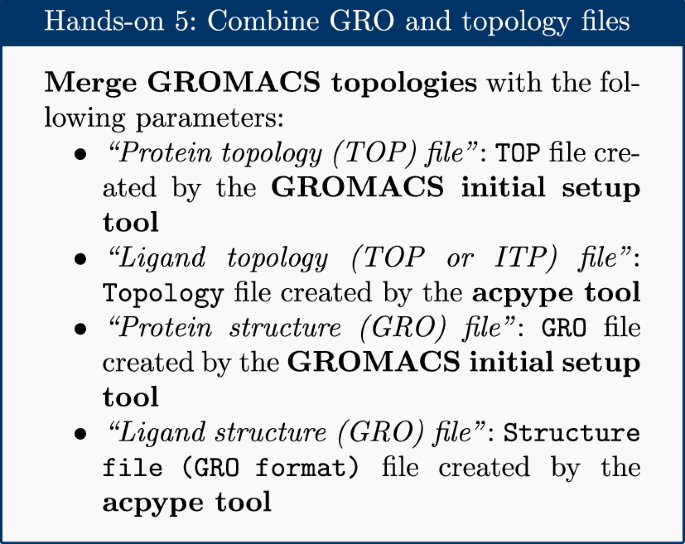
Intuitive, reproducible high-throughput molecular dynamics in Galaxy: a tutorial | Journal of Cheminformatics | Full Text

Data including GROMACS input files for atomistic molecular dynamics simulations of mixed, asymmetric bilayers including molecular topologies, equilibrated structures, and force field for lipids compatible with OPLS-AA parameters - ScienceDirect

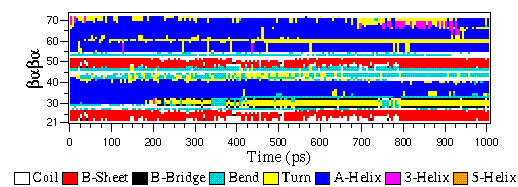




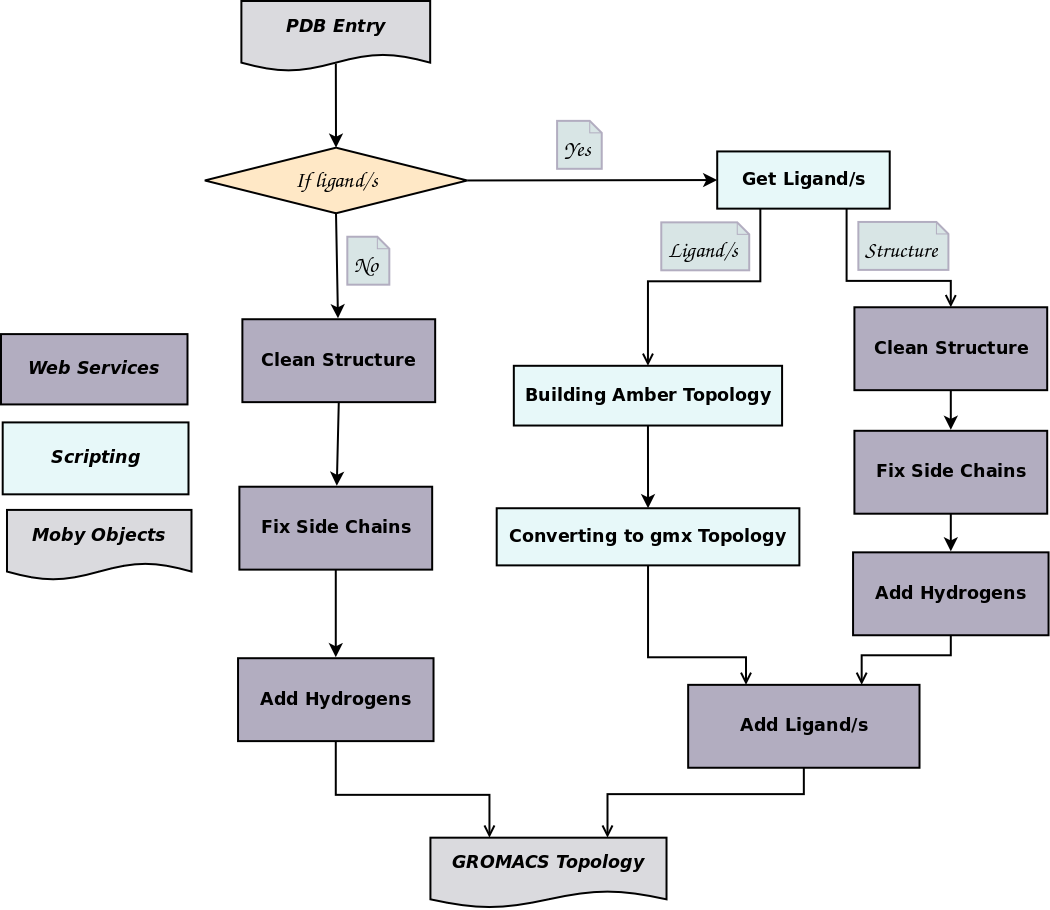

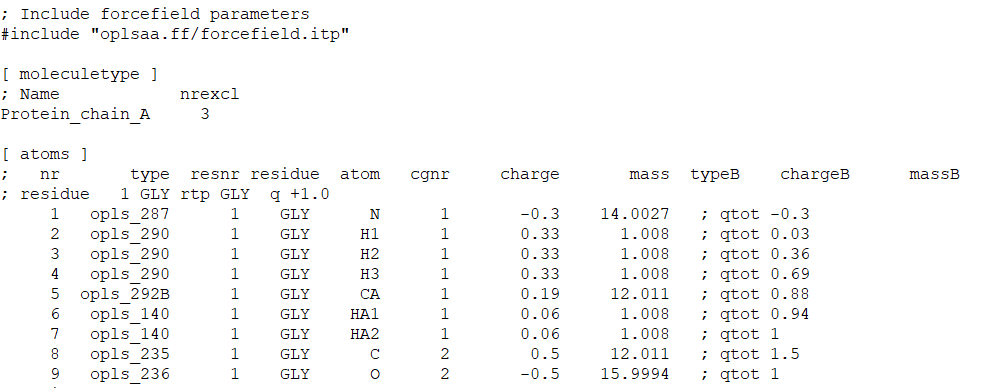


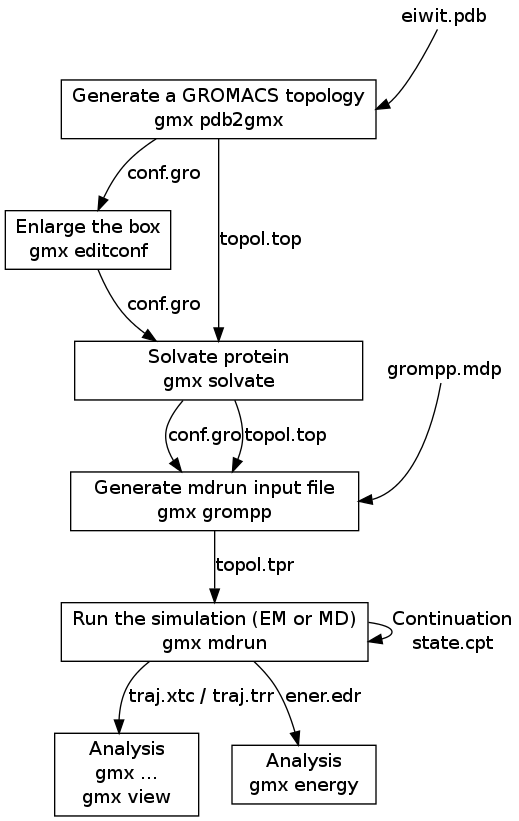

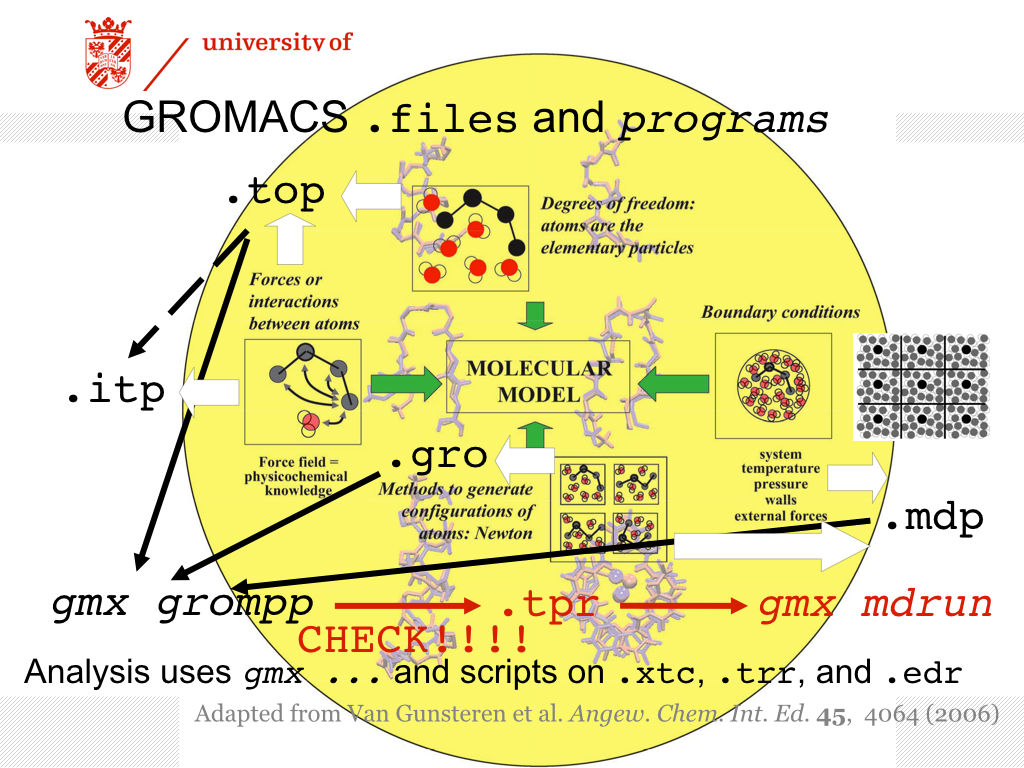


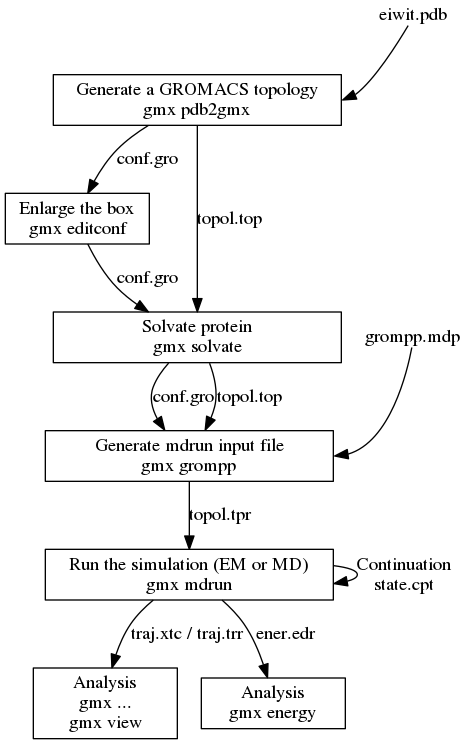



![PDF] GROMACS: A message-passing parallel molecular dynamics implementation | Semantic Scholar PDF] GROMACS: A message-passing parallel molecular dynamics implementation | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/625ddfe164e3a25405672ecc1ca17d05353ff182/8-Table1-1.png)
